Abstract
Background:
Chromosome 7 (chr7) abnormalities are commonly seen in patients with acute myeloid leukemia (AML) or myelodysplastic syndrome and are associated with poor prognosis. Flow cytometry (FCM) is typically used clinically to quantify residual malignancy during treatment but the relationship of cell surface immunophenotype with genetic features is incompletely defined. Single-cell RNA sequencing (scRNA-seq) with oligonucleotide-conjugated antibodies may be able to integrate cytogenetic genotypes found within leukemic clones with specific transcriptomic and immunophenotypic signatures.
Methods:
Bone marrow (BM) aspirate was collected from 12 AML patients with known abnormalities on chr7 according to cytogenetics and 3 healthy donors (HDs). We previously established a reference for normal immune cells where we assessed the BM of 20 HDs (PMID:30518681). HDs in present study were selected from this same cohort. Multi-parameter FCM was performed as previously described and scRNA-seq with 31 antibodies (10x Genomics, 3'v2) was performed on BM mononuclear cells. Data from 16 samples (including 1 patient replicate) were mapped onto the atlas with dimension reduction in gene expression (GEX) by principal component analysis and Uniform Manifold Approximation and Projection (UMAP). Cell clusters were constructed and surface proteins were utilized to determine cellular annotation.
Patient GEX profiles were compared to those of HDs, and two algorithms were used to identify malignant cells with chromosomal abnormalities. Alterations of large chromosomal segments were identified by Hidden Markov Model. Clinical cytogenetics and expression matrices from the model output were combined to annotate individual cells as malignant or normal for all potential regions. Machine learning classifiers were applied to predict malignant cells using protein expression and important proteins were selected by models with measure for significant features.
Results:
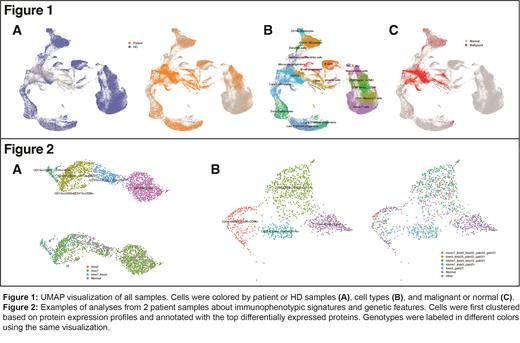
After quality control, 132,658 cells were included, of which 43,441 and 12,572 cells from AML patients and HD respectively had additional cell surface immunophenotyping data. Samples were well-integrated by UMAP (Figure 1A) and cell annotation overlapped with previously reported annotations by GEX of HDs (Figure 1B), with 15,270 malignant cells identified (Figure 1C). Malignant groups were created if more than 20 cells shared the same chromosomal alterations. Among all 12 patients, 3 of them only had monosomy 7 as the sole genetic aberration, while 9 patients had abnormalities on other chromosomes.
The correlation between cell types showed distinct features among patients, and cells in different groups had different protein expression profiles. For example, 22% of the cells (n = 432) from the myeloid population of patient 1 (n = 1957) were identified as normal and 77% had a loss of chr7 (Figure 2A). Clustering in lower resolution separated them into two groups: CD11b+CD33+ (n = 1026) and CD133+CD34+ (n = 931). The CD133+CD34+ group had a higher percentage of malignant cells overall (74% vs. 83%) while a subset of the CD11b+CD33+ group (CD16+CD13+) had the highest percentage of cells with losses on both chr2 and chr7 (60%). Data from patient 2 also suggested that cells with different immunophenotypes were estimated to have different chromosomal changes (Figure 2B). While 69% of the malignant cells had a loss on chr7, most of those without changes on chr7 were from CD45+CD11b+ group where 62% of the cells did not have a detectable change on chr7.
The top significant proteins for distinguishing malignant from normal cells among all patients were CD117, CD33, CD34, CD44, and CD47. FCM intensity was compared with the scRNA-seq immunophenotyping data, confirming the similarity in distribution. The scRNA-seq data with immunophenotyping can capture and recapitulate the leukemic immunophenotype, further linking it with copy number changes to reveal potential subclonal structures.
Conclusion:
By comparing the expression profile of AML patients with abnormalities on chr7 to HDs, this study provides evidence that leukemic immunophenotype is correlated with chromosomal structural changes. The experiments on a single-cell level were able to identify clones in higher resolution and revealed potential cell surface protein markers that could be used clinically to identify specific malignant populations in patients with myeloid malignancies.
Ghiaur: Syros Pharmaceuticals: Consultancy; Menarini Richerche: Research Funding. Hourigan: Sellas: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal